A Note on bmlm’s Spaghetti Plots
Matti Vuorre
2023-09-26
Source:vignettes/Spaghetti-Plots/Spaghetti-Plots.Rmd
Spaghetti-Plots.RmdConsider a mediation model in which Y is binary:
## # A tibble: 6 × 5
## id x m y biny
## <int> <dbl> <dbl> <dbl> <int>
## 1 101 0.333 0.983 -1.44 0
## 2 101 0.333 0.928 0.144 0
## 3 101 0.333 -0.719 -1.63 0
## 4 101 1.33 0.745 1.92 1
## 5 101 -1.67 -0.124 -1.93 0
## 6 101 -0.667 -1.27 0.688 1fit_biny <- mlm(d = BLch9_biny,
id = "id",
y = "biny", binary_y = TRUE,
cores=4, iter = 500)## Warning: The largest R-hat is NA, indicating chains have not mixed.
## Running the chains for more iterations may help. See
## https://mc-stan.org/misc/warnings.html#r-hat## Warning: Bulk Effective Samples Size (ESS) is too low, indicating posterior means and medians may be unreliable.
## Running the chains for more iterations may help. See
## https://mc-stan.org/misc/warnings.html#bulk-ess## Warning: Tail Effective Samples Size (ESS) is too low, indicating posterior variances and tail quantiles may be unreliable.
## Running the chains for more iterations may help. See
## https://mc-stan.org/misc/warnings.html#tail-essmlm_summary(fit_biny) %>%
knitr::kable()| Parameter | Mean | SE | Median | 2.5% | 97.5% | n_eff | Rhat |
|---|---|---|---|---|---|---|---|
| a | 0.19 | 0.04 | 0.19 | 0.12 | 0.26 | 775 | 1.00 |
| b | 0.26 | 0.05 | 0.26 | 0.16 | 0.37 | 630 | 1.01 |
| cp | 0.24 | 0.05 | 0.24 | 0.14 | 0.34 | 696 | 1.00 |
| me | 0.10 | 0.03 | 0.10 | 0.05 | 0.15 | 346 | 1.01 |
| c | 0.34 | 0.06 | 0.33 | 0.22 | 0.45 | 660 | 1.01 |
| pme | 0.30 | 0.08 | 0.29 | 0.17 | 0.46 | 398 | 1.01 |
The mlm_spaghetti_plot() function produces figures in
which the horizontal axis values of the M-Y graph (right panel, below)
correspond to the vertical axis values of the X-M graph (left panel,
below):
pasta <- mlm_spaghetti_plot(fit_biny, BLch9_biny,
id="id", x="x", m="m", y="biny",
binary_y=T)
gridExtra::grid.arrange(
pasta[[1]],
pasta[[2]] + coord_cartesian(ylim = 0:1),
nrow = 1)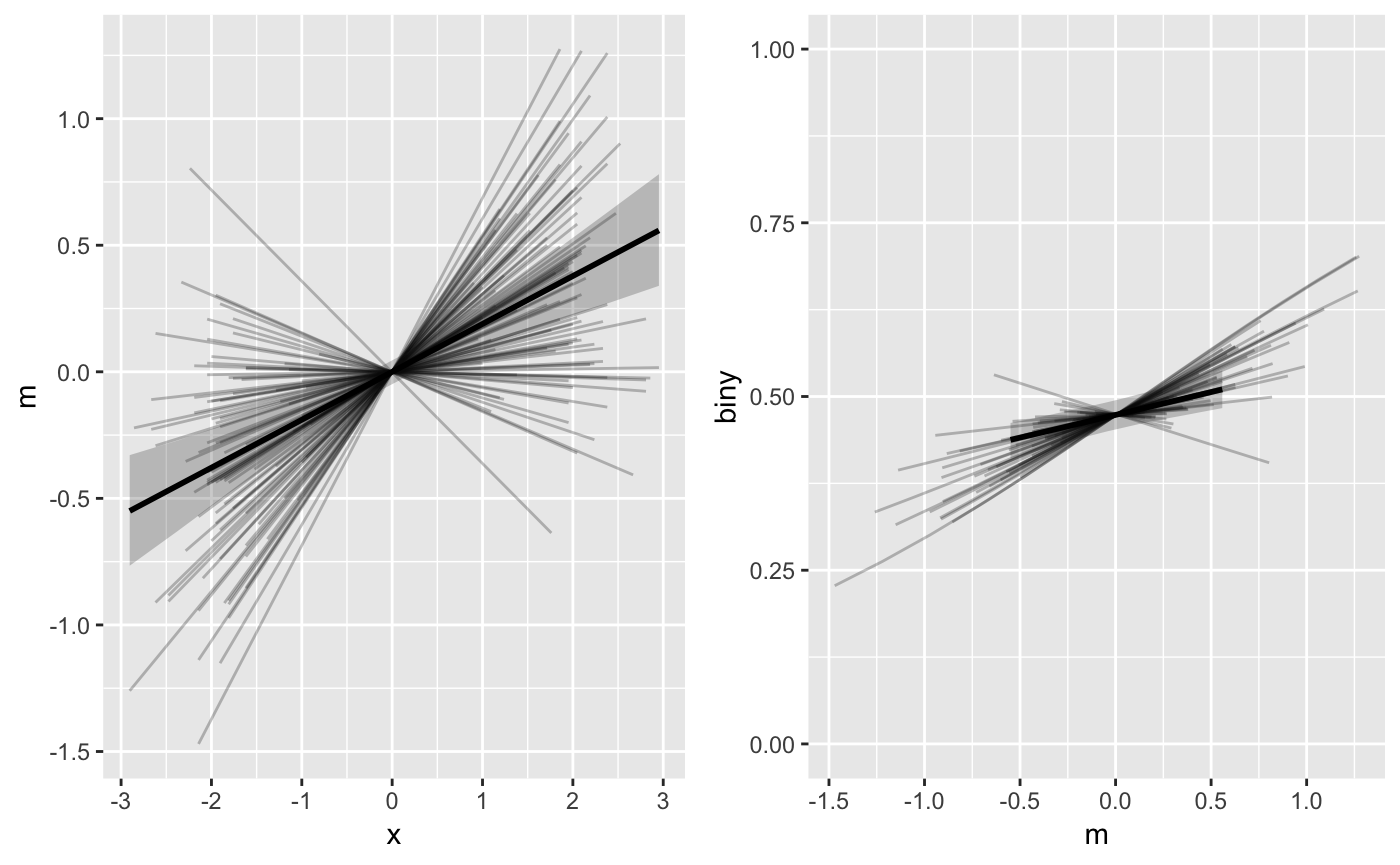
It is important to note that the fitted values (lines) on the right
panel of this figure are therefore based on the variation of the fitted
values in the left panel, and not on the observed values of
m. This behavior usually makes sense and is the default
option for mlm_spaghetti_plot(). However, sometimes users
may want to plot the M-Y relationship’s (b path’s) fitted values with
the observed data values of M on the horizontal axis instead.
To do this, set mx = "data":
pasta2 <- mlm_spaghetti_plot(fit_biny, BLch9_biny,
id="id", x="x", m="m", y="biny",
binary_y=T, mx = "data")
gridExtra::grid.arrange(
pasta2[[1]],
pasta2[[2]] + coord_cartesian(ylim = 0:1),
nrow = 1)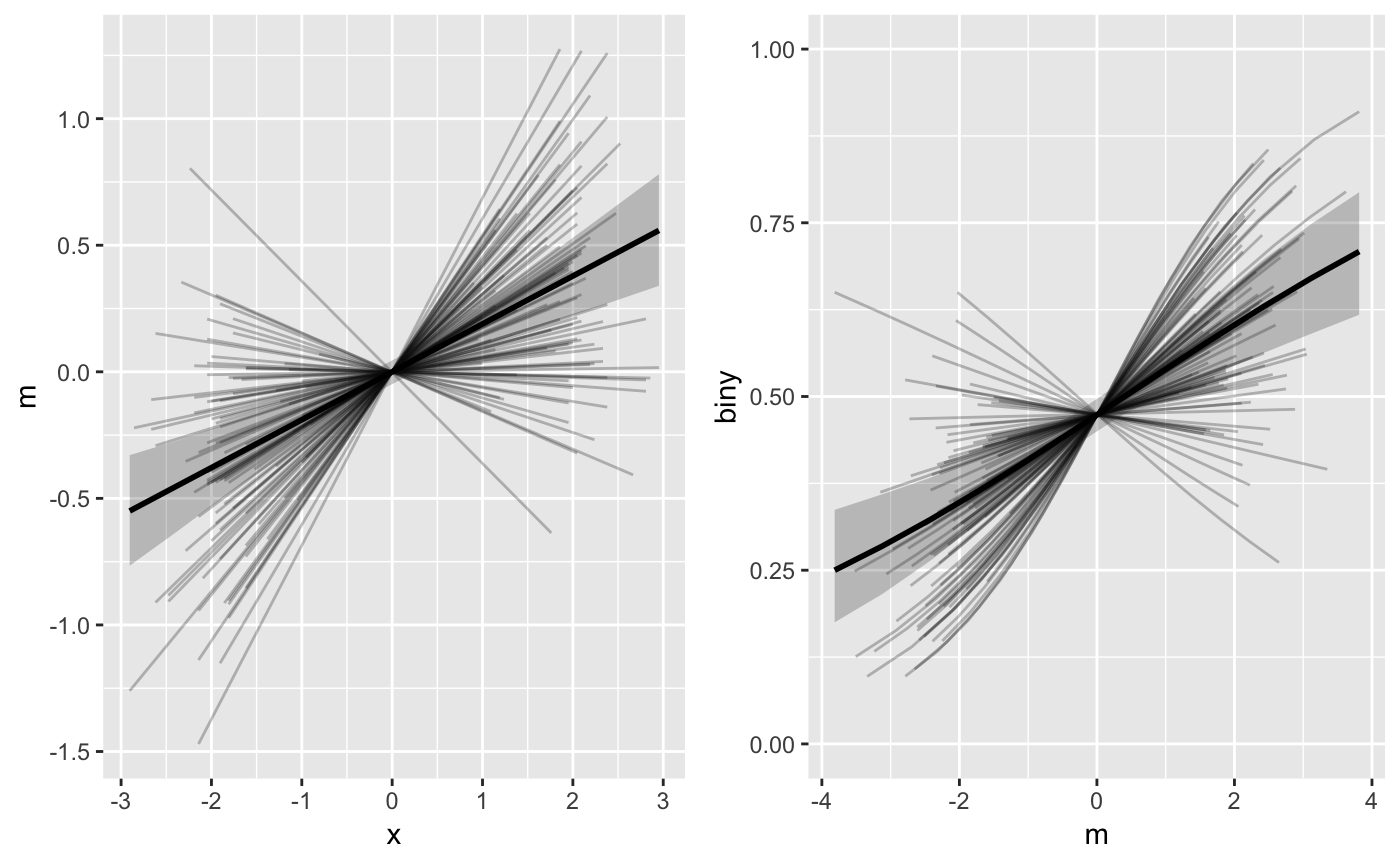
As result, the horizontal axis variability for the b path figure (right panel) is much greater. However, the two panels are no longer directly comparable (vertical axis in left panel doesn’t match horizontal axis of right panel) or interpretable as the model’s implied mediated effect path.
Thanks for reading. Please notify the package’s developer on GitHub if you have further questions or comments.