Data Analysis for V&M (2017)
Matti Vuorre
2017-05-01
library(knitr)
library(gridExtra)
library(lme4)
library(tidyverse)
library(brms)
library(vmapp2017)
knitr::opts_chunk$set(message = F)Functions and constants
get_pse <- function(X, x, c, scale = 1) {
# Function to get PSEs
# X = Matrix of posterior samples (or row of MLEs)
# x = condition (0 or 1)
# c = constant (what value ISI is centered on)
pse <- - ( (X[,1] + X[,3]*x) / (X[,2] + X[,4]*x) )
pse <- (scale * pse) + c
return(pse)
}
iters <- 17500
warmups <- 5000
color_vals = c("#e41a1c", "#377eb8")
alphas <- .2
# Estimated models are saved in a local file to save time
load(file="../../../~$models.rda")Use common plotting theme
library(vmisc)
theme_set(theme_pub())Data preparation
The data is contained in this package. Here we collapse the illusion data to counts (models estimate faster with collapsed data)
data(illusion)
illusion <- illusion %>%
group_by(exp, id, condition, cond, interval) %>%
summarise(k = sum(response, na.rm=T),
n = n(),
exclude = unique(exclude)) %>%
ungroup()Experiment 1
Apparent Motion
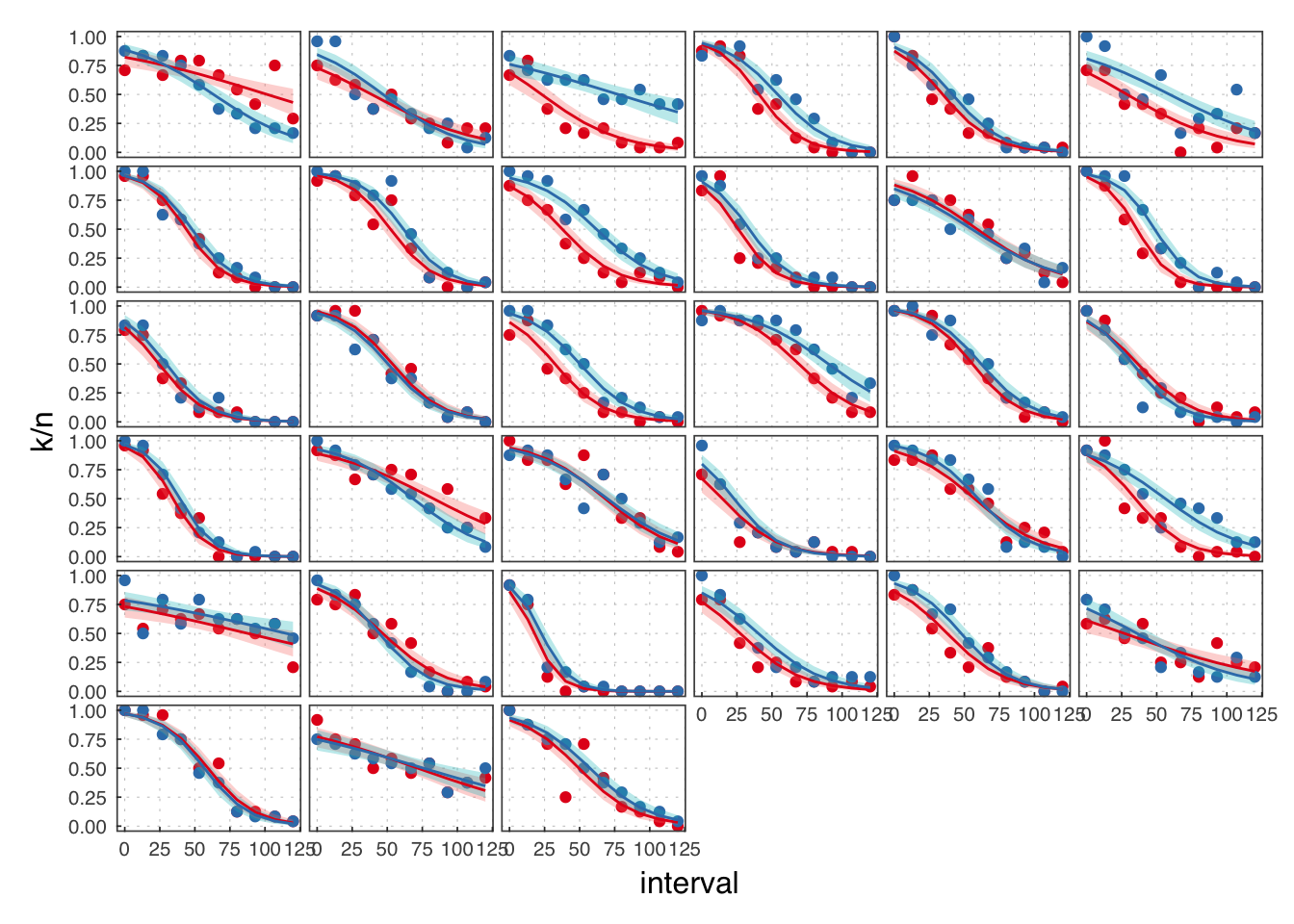
( exp1 <- filter(illusion, exp == 1) )## # A tibble: 384 × 8
## exp id condition cond interval k n exclude
## <chr> <dbl> <chr> <dbl> <dbl> <dbl> <int> <lgl>
## 1 1 101 involuntary 0 33 9 20 FALSE
## 2 1 101 involuntary 0 50 8 20 FALSE
## 3 1 101 involuntary 0 83 8 20 FALSE
## 4 1 101 involuntary 0 100 6 20 FALSE
## 5 1 101 involuntary 0 133 6 20 FALSE
## 6 1 101 involuntary 0 150 5 20 FALSE
## 7 1 101 involuntary 0 200 4 20 FALSE
## 8 1 101 involuntary 0 300 4 20 FALSE
## 9 1 101 voluntary 1 33 10 20 FALSE
## 10 1 101 voluntary 1 50 10 20 FALSE
## # ... with 374 more rows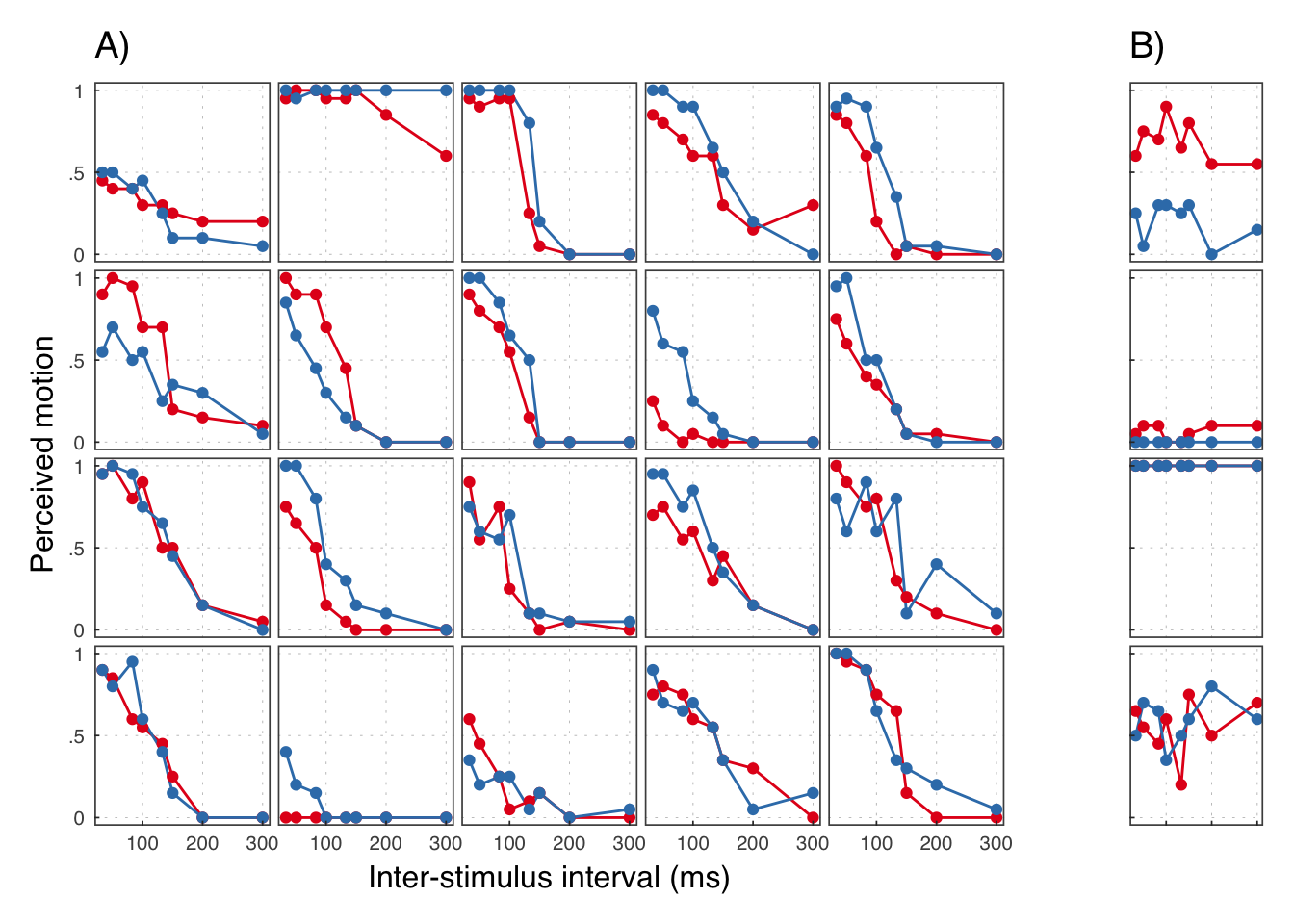
We reject the participants in B.
exp1 <- filter(exp1, exclude == F)
Cc <- mean(unique(exp1$interval))
exp1$isic <- ( exp1$interval - Cc ) / 10GLMM MLE estimation
exp1mle <- glmer(k/n ~ isic*condition + (isic*condition|id),
weights = n, family=binomial(link=logit),
data=exp1)summary(exp1mle)## Generalized linear mixed model fit by maximum likelihood (Laplace
## Approximation) [glmerMod]
## Family: binomial ( logit )
## Formula: k/n ~ isic * condition + (isic * condition | id)
## Data: exp1
## Weights: n
##
## AIC BIC logLik deviance df.resid
## 1256.0 1308.8 -614.0 1228.0 306
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.47676 -0.57691 -0.07241 0.63698 2.81210
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## id (Intercept) 3.32248 1.8228
## isic 0.01843 0.1358 0.07
## conditionvoluntary 0.51110 0.7149 -0.47 -0.23
## isic:conditionvoluntary 0.02167 0.1472 0.49 -0.24 -0.47
## Number of obs: 320, groups: id, 20
##
## Fixed effects:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -1.06610 0.41767 -2.553 0.0107 *
## isic -0.29819 0.03363 -8.868 <2e-16 ***
## conditionvoluntary 0.48399 0.19325 2.504 0.0123 *
## isic:conditionvoluntary -0.01918 0.03860 -0.497 0.6192
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) isic cndtnv
## isic 0.121
## cndtnvlntry -0.475 -0.281
## isc:cndtnvl 0.363 -0.340 -0.189
## convergence code: 0
## Model failed to converge with max|grad| = 0.00229299 (tol = 0.001, component 1)GLMM Bayes estimation
Priors <- c(
set_prior("normal(0, 100)", class = "b"),
set_prior("cauchy(0, 4)", class = "sd")
)exp1b <- brm(k | trials(n) ~ isic*condition + (isic*condition|id),
family=binomial(link=logit), prior = Priors,
data=exp1, cores = 4, iter = iters, warmup = warmups)## Inference for Stan model: binomial(logit) brms-model.
## 4 chains, each with iter=17500; warmup=5000; thin=1;
## post-warmup draws per chain=12500, total post-warmup draws=50000.
##
## mean se_mean sd 2.5% 97.5% n_eff Rhat
## b_Intercept -1.08 0.01 0.49 -2.08 -0.14 8825 1
## b_isic -0.30 0.00 0.04 -0.38 -0.22 17651 1
## b_conditionvoluntary 0.51 0.00 0.23 0.07 0.99 21705 1
## b_isic:conditionvoluntary -0.02 0.00 0.05 -0.12 0.07 22646 1
##
## Samples were drawn using NUTS(diag_e) at Thu Feb 23 10:40:36 2017.
## For each parameter, n_eff is a crude measure of effective sample size,
## and Rhat is the potential scale reduction factor on split chains (at
## convergence, Rhat=1).# Posterior probability (one-sided p-value)
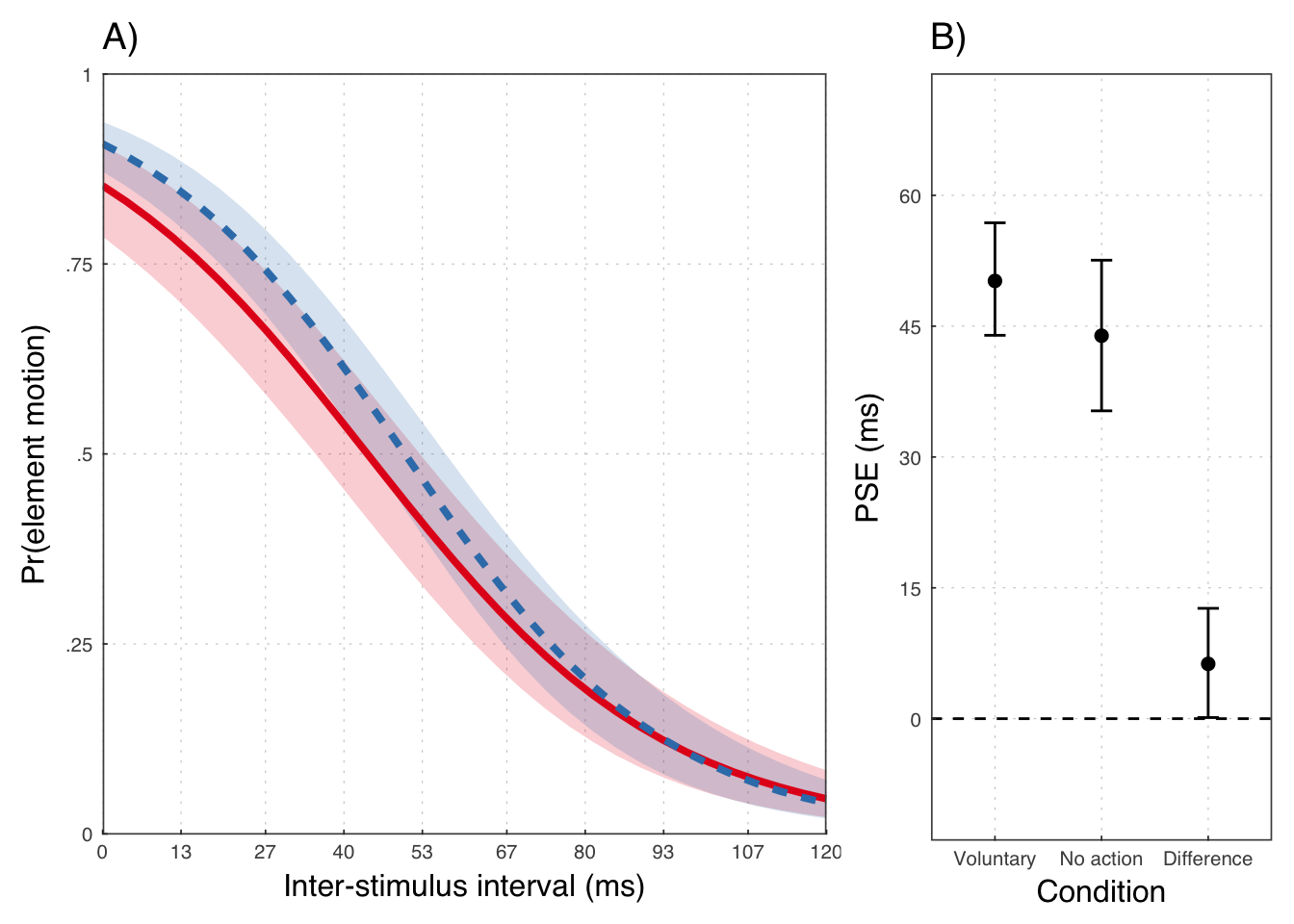
mean(posterior_samples(exp1b, pars = "b_cond") > 0) * 100.0## [1] 98.758Population level PSEs
## # A tibble: 3 × 4
## key m lwr upr
## <chr> <dbl> <dbl> <dbl>
## 1 Difference 18.73404 1.809483 37.78756
## 2 Involuntary 94.67529 59.582235 126.39876
## 3 Voluntary 113.40933 84.382228 142.05608## # A tibble: 3 × 2
## key pp
## <chr> <dbl>
## 1 Difference 0.98334
## 2 Involuntary 0.99986
## 3 Voluntary 1.00000Plot results
e1p4 <- ggplot(exp1psesum, aes(x=reorder(key, rev(m)), m)) +
geom_hline(yintercept = 0, lty=2) +
geom_point(size=2) +
geom_errorbar(aes(ymin=lwr, ymax=upr), width = .2, size=.5) +
scale_y_continuous("PSE (ms)",
breaks = c(-30, 0, 30, 60, 90, 120, 150),
limits = c(-35, 155)) +
scale_x_discrete("Condition",
labels = c("Voluntary", "No action", "Difference"))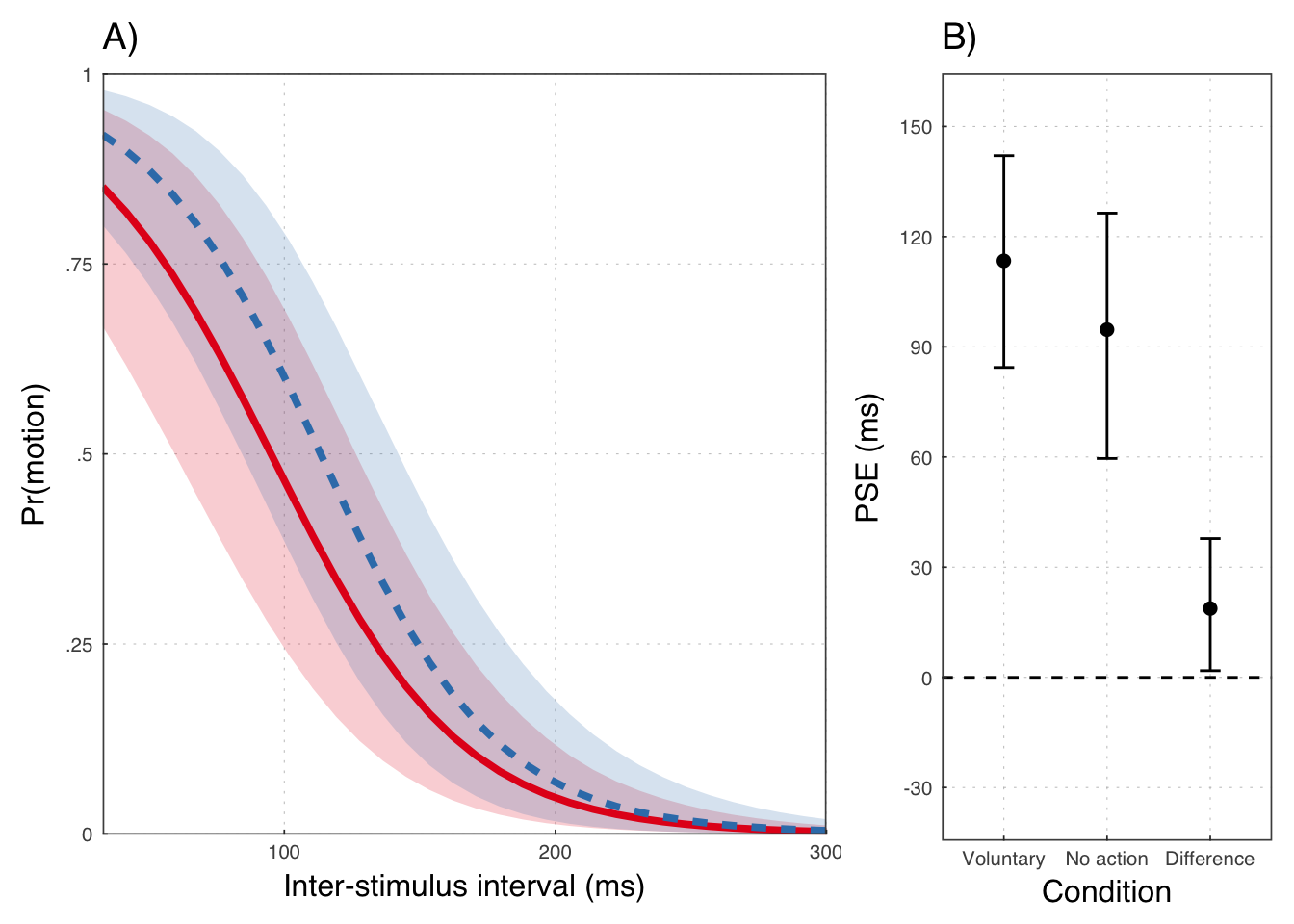
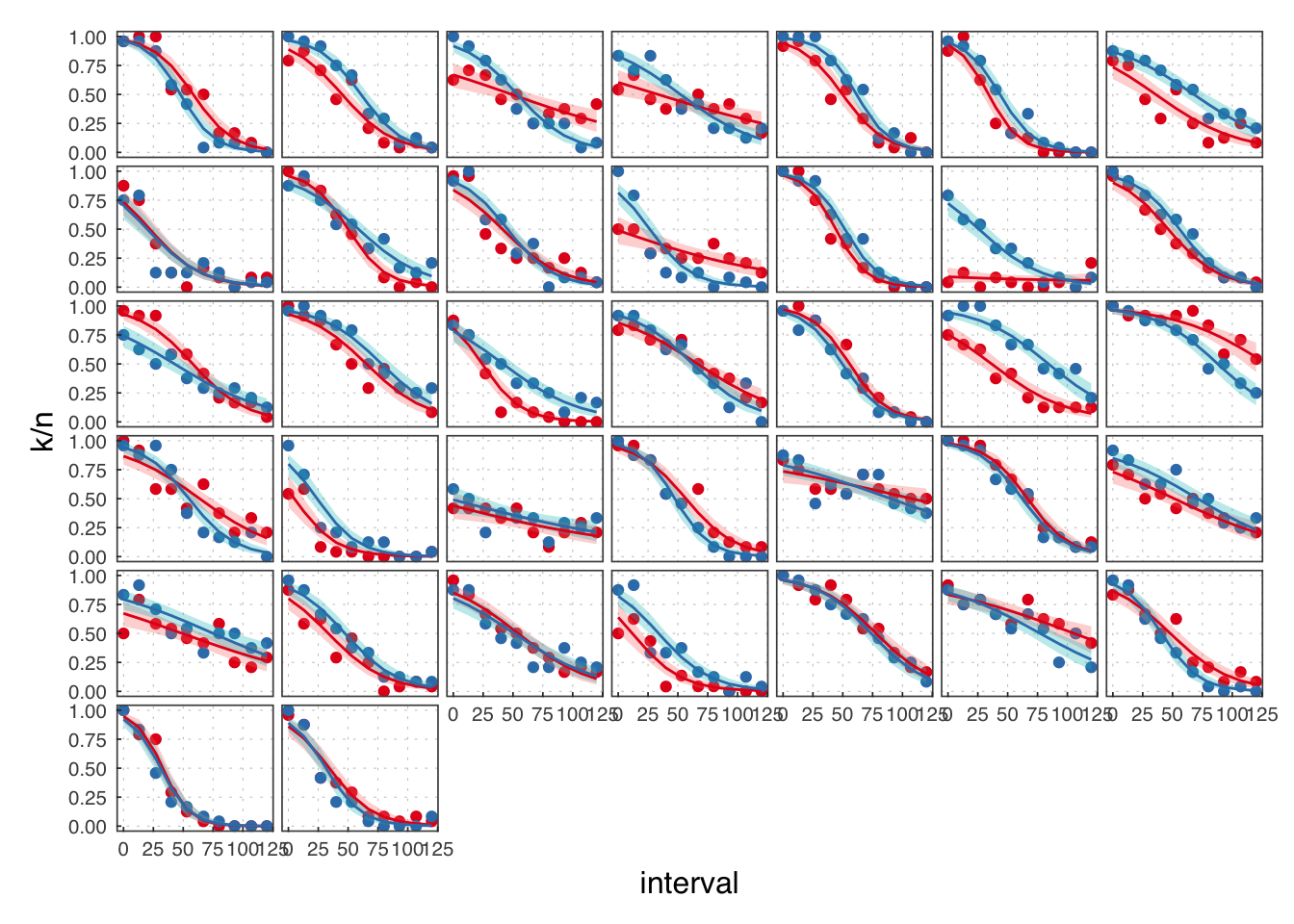
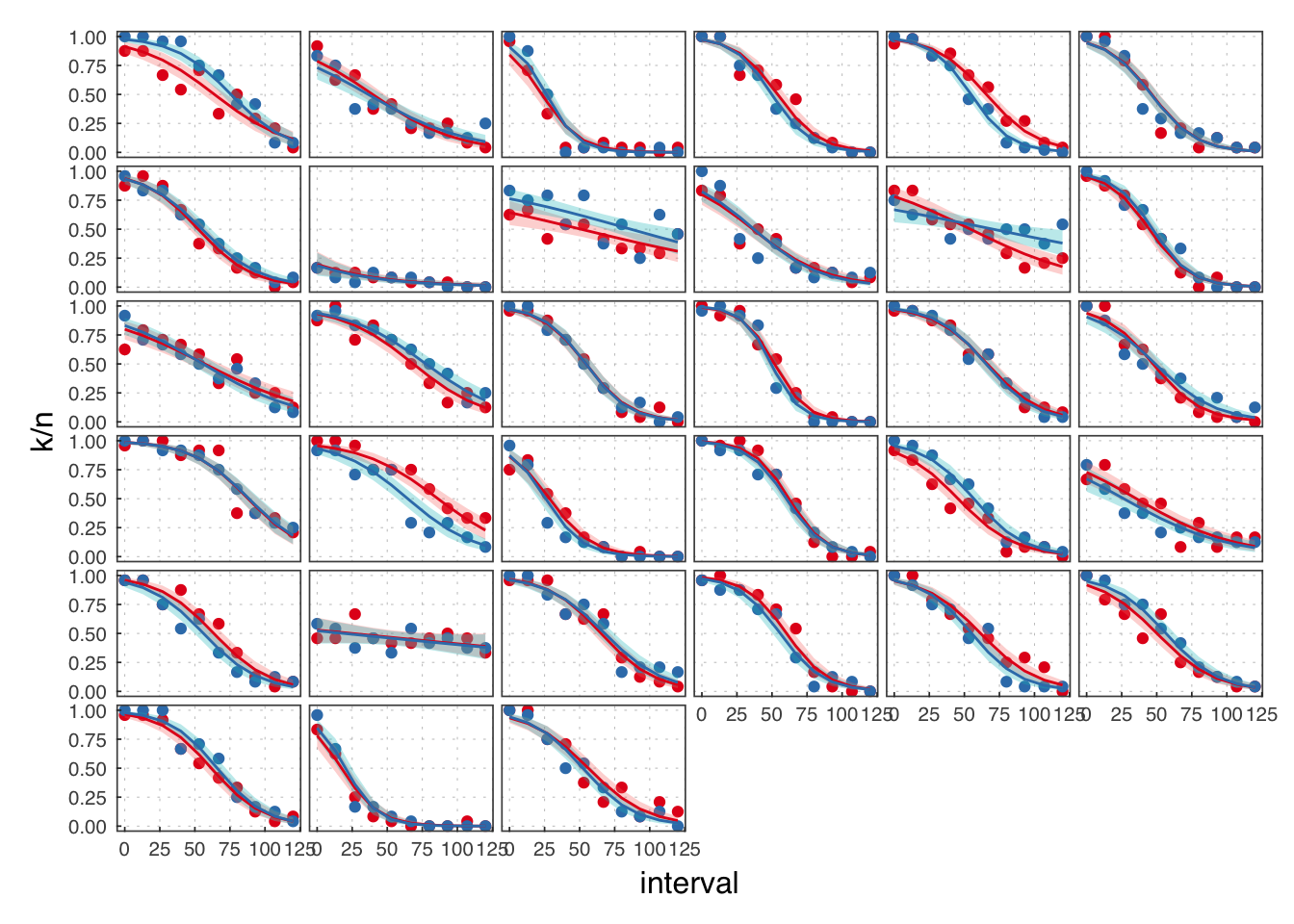
Throughout, we use red to refer to the passive observation condition, and blue to the voluntary action condition.

Interval Estimation
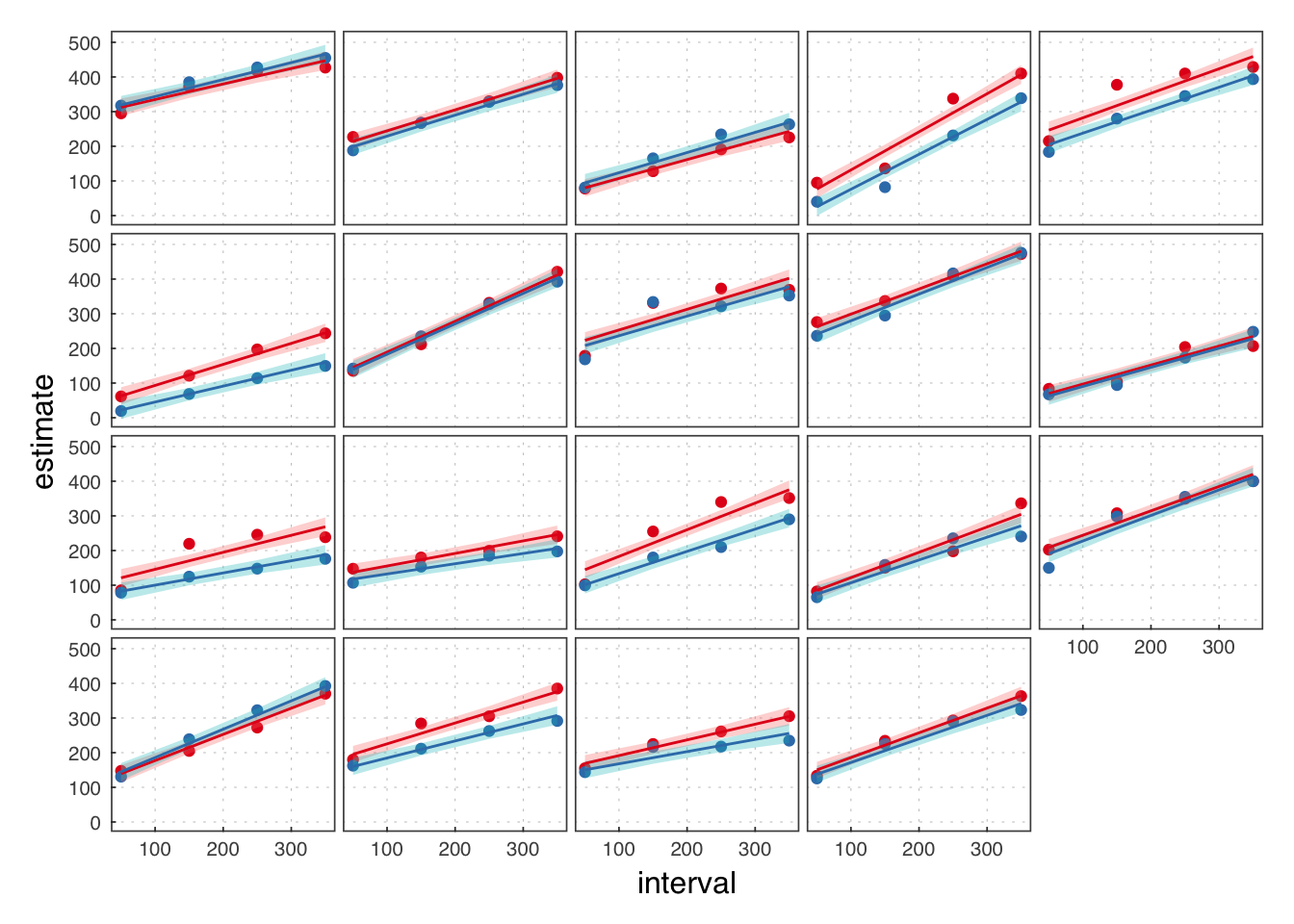
data(ie)
ie$isic <- ( ie$interval - mean(unique(ie$interval)) ) / 10
ie## # A tibble: 3,011 × 6
## id condition interval estimate cond isic
## <dbl> <chr> <int> <dbl> <dbl> <dbl>
## 1 101 voluntary 350 500 1 15
## 2 101 voluntary 250 500 1 5
## 3 101 voluntary 150 400 1 -5
## 4 101 voluntary 150 450 1 -5
## 5 101 voluntary 350 500 1 15
## 6 101 voluntary 250 500 1 5
## 7 101 voluntary 250 450 1 5
## 8 101 voluntary 350 450 1 15
## 9 101 voluntary 50 350 1 -15
## 10 101 voluntary 50 350 1 -15
## # ... with 3,001 more rowsLMM MLE
iemle <- lmerTest::lmer(estimate ~ isic*condition + (isic*condition|id),
data = ie)summary(iemle)## Linear mixed model fit by REML ['merModLmerTest']
## Formula: estimate ~ isic * condition + (isic * condition | id)
## Data: ie
##
## REML criterion at convergence: 35132.8
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -5.0022 -0.5597 -0.0718 0.4881 5.3010
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## id (Intercept) 5110.4748 71.4876
## isic 3.3282 1.8243 0.14
## conditionvoluntary 855.9967 29.2574 0.10 -0.28
## isic:conditionvoluntary 0.9201 0.9592 0.35 -0.13 0.96
## Residual 6492.0292 80.5731
## Number of obs: 3011, groups: id, 19
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 257.7719 16.5318 15.593
## isic 6.5033 0.4580 14.200
## conditionvoluntary -25.3937 7.3266 -3.466
## isic:conditionvoluntary -0.3991 0.3428 -1.164
##
## Correlation of Fixed Effects:
## (Intr) isic cndtnv
## isic 0.131
## cndtnvlntry 0.060 -0.234
## isc:cndtnvl 0.223 -0.295 0.567LMM Bayes
ieb <- brm(estimate ~ isic*condition + (isic*condition|id),
data=ie, cores = 4, iter = iters, warmup = warmups)## Inference for Stan model: gaussian(identity) brms-model.
## 4 chains, each with iter=17500; warmup=5000; thin=1;
## post-warmup draws per chain=12500, total post-warmup draws=50000.
##
## mean se_mean sd 2.5% 97.5% n_eff Rhat
## b_Intercept 257.78 0.21 19.01 220.03 295.23 8164 1
## b_isic 6.50 0.00 0.52 5.47 7.55 18232 1
## b_conditionvoluntary -25.39 0.05 8.07 -41.37 -9.37 23309 1
## b_isic:conditionvoluntary -0.40 0.00 0.36 -1.10 0.30 50000 1
##
## Samples were drawn using NUTS(diag_e) at Thu Feb 23 10:49:21 2017.
## For each parameter, n_eff is a crude measure of effective sample size,
## and Rhat is the potential scale reduction factor on split chains (at
## convergence, Rhat=1).# Posterior probability (one-sided p-value)
mean(posterior_samples(ieb, pars = "b_cond") < 0) * 100## [1] 99.794Plot results
new_int <- sort(unique(ie$isic))
iepost <- posterior_samples(ieb, pars=parnames(ieb)[1:4])
diffdat <- expand.grid(intrcpt = 0,
isic = 0,
condition = 1,
intrct = new_int)
# create predictor matrix
Xmat <- as.matrix(diffdat)
# matrix for fitted values
fitmat <- matrix(ncol=nrow(iepost), nrow=nrow(diffdat))
# obtain predicted values for each row
for (i in 1:nrow(iepost)) {
fitmat[,i] <- Xmat %*% as.matrix(iepost)[i,]
}
# get quantiles to plot
diffdat$lower <- apply(fitmat, 1, quantile, prob=0.025)
diffdat$med <- apply(fitmat, 1, quantile, prob=0.5)
diffdat$upper <- apply(fitmat, 1, quantile, prob=0.975)
diffdat$variable <- sort(unique(ie$isic))
e1bp6 <- ggplot(diffdat, aes(x = variable, y=med)) +
geom_hline(yintercept = 0, lty=2) +
geom_point(size = 1.4) +
geom_errorbar(aes(ymin=lower, ymax=upper), width = 1.3, size=.3) +
scale_y_continuous("Difference (ms)",
breaks = c(20, 0, -20, -40, -60, -80),
limits = c(-85, 25)) +
scale_x_continuous("ISI (ms)",
breaks = c(-15, -5, 5, 15),
labels = c("50", "150", "250", "350"),
limits = c(-18, 18))e1bp7 <- grid.arrange(
e1bp5 +
ggtitle("A)"),
e1bp6 +
ggtitle("B)"),
widths = c(7, 3))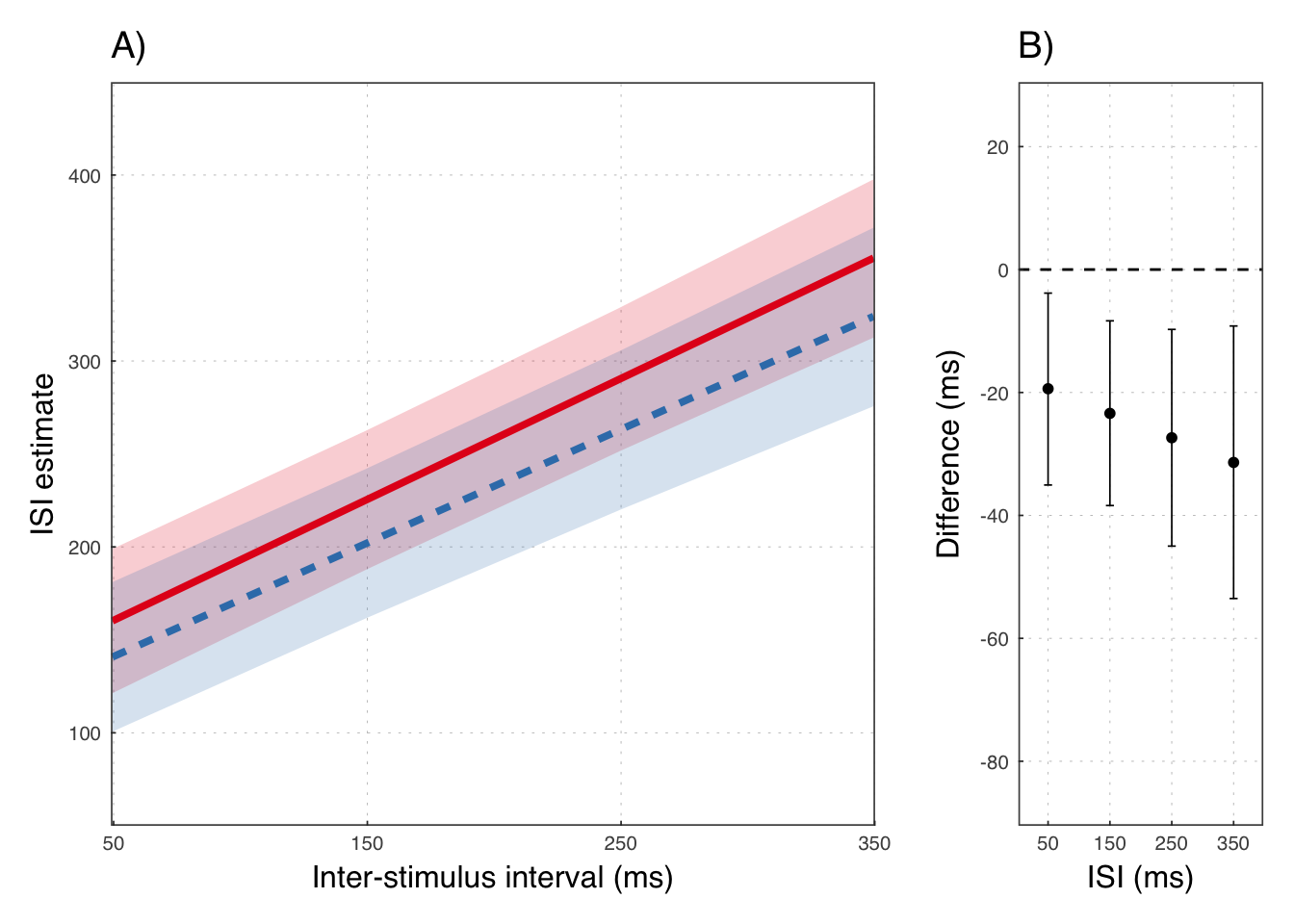
Experiment 2
( exp2 <- filter(illusion, exp == 2) )## # A tibble: 720 × 8
## exp id condition cond interval k n exclude
## <chr> <dbl> <chr> <dbl> <dbl> <dbl> <int> <lgl>
## 1 2 202 involuntary 0 0 17 24 FALSE
## 2 2 202 involuntary 0 13 20 24 FALSE
## 3 2 202 involuntary 0 27 16 24 FALSE
## 4 2 202 involuntary 0 40 19 24 FALSE
## 5 2 202 involuntary 0 53 19 24 FALSE
## 6 2 202 involuntary 0 67 16 24 FALSE
## 7 2 202 involuntary 0 80 13 24 FALSE
## 8 2 202 involuntary 0 93 10 24 FALSE
## 9 2 202 involuntary 0 107 18 24 FALSE
## 10 2 202 involuntary 0 120 7 24 FALSE
## # ... with 710 more rows
We reject the participants in B.
exp2 <- filter(exp2, exclude == F)
Cc <- 50 # ISI to center on (around middle)
exp2$isic <- ( exp2$interval - Cc ) / 10GLMM MLE estimation
exp2mle <- glmer(k/n ~ isic*condition + (isic*condition|id),
weights=n, family=binomial(link=logit),
data=exp2)summary(exp2mle)## Generalized linear mixed model fit by maximum likelihood (Laplace
## Approximation) [glmerMod]
## Family: binomial ( logit )
## Formula: k/n ~ isic * condition + (isic * condition | id)
## Data: exp2
## Weights: n
##
## AIC BIC logLik deviance df.resid
## 2945.7 3008.6 -1458.8 2917.7 646
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -3.6524 -0.6673 -0.0274 0.7371 3.5086
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## id (Intercept) 0.91930 0.9588
## isic 0.05073 0.2252 0.64
## conditionvoluntary 0.26396 0.5138 -0.53 -0.41
## isic:conditionvoluntary 0.01289 0.1135 -0.29 -0.45 0.68
## Number of obs: 660, groups: id, 33
##
## Fixed effects:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -0.44381 0.17103 -2.595 0.009462 **
## isic -0.50168 0.04127 -12.156 < 2e-16 ***
## conditionvoluntary 0.38996 0.10243 3.807 0.000141 ***
## isic:conditionvoluntary 0.01168 0.02630 0.444 0.656854
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) isic cndtnv
## isic 0.618
## cndtnvlntry -0.527 -0.374
## isc:cndtnvl -0.248 -0.474 0.514GLMM Bayes estimation
exp2b <- brm(k | trials(n) ~ isic*condition + (isic*condition|id),
family=binomial(link=logit), prior = Priors,
data=exp2, cores = 4, iter = iters, warmup = warmups)## Inference for Stan model: binomial(logit) brms-model.
## 4 chains, each with iter=17500; warmup=5000; thin=1;
## post-warmup draws per chain=12500, total post-warmup draws=50000.
##
## mean se_mean sd 2.5% 97.5% n_eff Rhat
## b_Intercept -0.44 0 0.18 -0.79 -0.08 7192 1
## b_isic -0.50 0 0.04 -0.59 -0.42 10762 1
## b_conditionvoluntary 0.38 0 0.11 0.17 0.60 16949 1
## b_isic:conditionvoluntary 0.01 0 0.03 -0.05 0.06 27036 1
##
## Samples were drawn using NUTS(diag_e) at Thu Feb 23 10:53:29 2017.
## For each parameter, n_eff is a crude measure of effective sample size,
## and Rhat is the potential scale reduction factor on split chains (at
## convergence, Rhat=1).# Posterior probability (one-sided p-value)
mean(posterior_samples(exp2b, pars = "b_cond") > 0) * 100## [1] 99.936Population-level PSEs.
## # A tibble: 3 × 4
## key m lwr upr
## <chr> <dbl> <dbl> <dbl>
## 1 Difference 7.669065 3.575212 11.93039
## 2 Involuntary 41.370815 35.199276 48.22684
## 3 Voluntary 49.039880 42.937926 56.04192## # A tibble: 3 × 2
## key pp
## <chr> <dbl>
## 1 Difference 0.9995
## 2 Involuntary 1.0000
## 3 Voluntary 1.0000Experiment 3
( exp3 <- filter(illusion, exp == 3) )## # A tibble: 760 × 8
## exp id condition cond interval k n exclude
## <chr> <dbl> <chr> <dbl> <dbl> <dbl> <int> <lgl>
## 1 3 302 involuntary 0 0 23 24 FALSE
## 2 3 302 involuntary 0 13 24 24 FALSE
## 3 3 302 involuntary 0 27 24 24 FALSE
## 4 3 302 involuntary 0 40 13 24 FALSE
## 5 3 302 involuntary 0 53 13 24 FALSE
## 6 3 302 involuntary 0 67 12 24 FALSE
## 7 3 302 involuntary 0 80 4 24 FALSE
## 8 3 302 involuntary 0 93 4 24 FALSE
## 9 3 302 involuntary 0 107 2 24 FALSE
## 10 3 302 involuntary 0 120 0 24 FALSE
## # ... with 750 more rows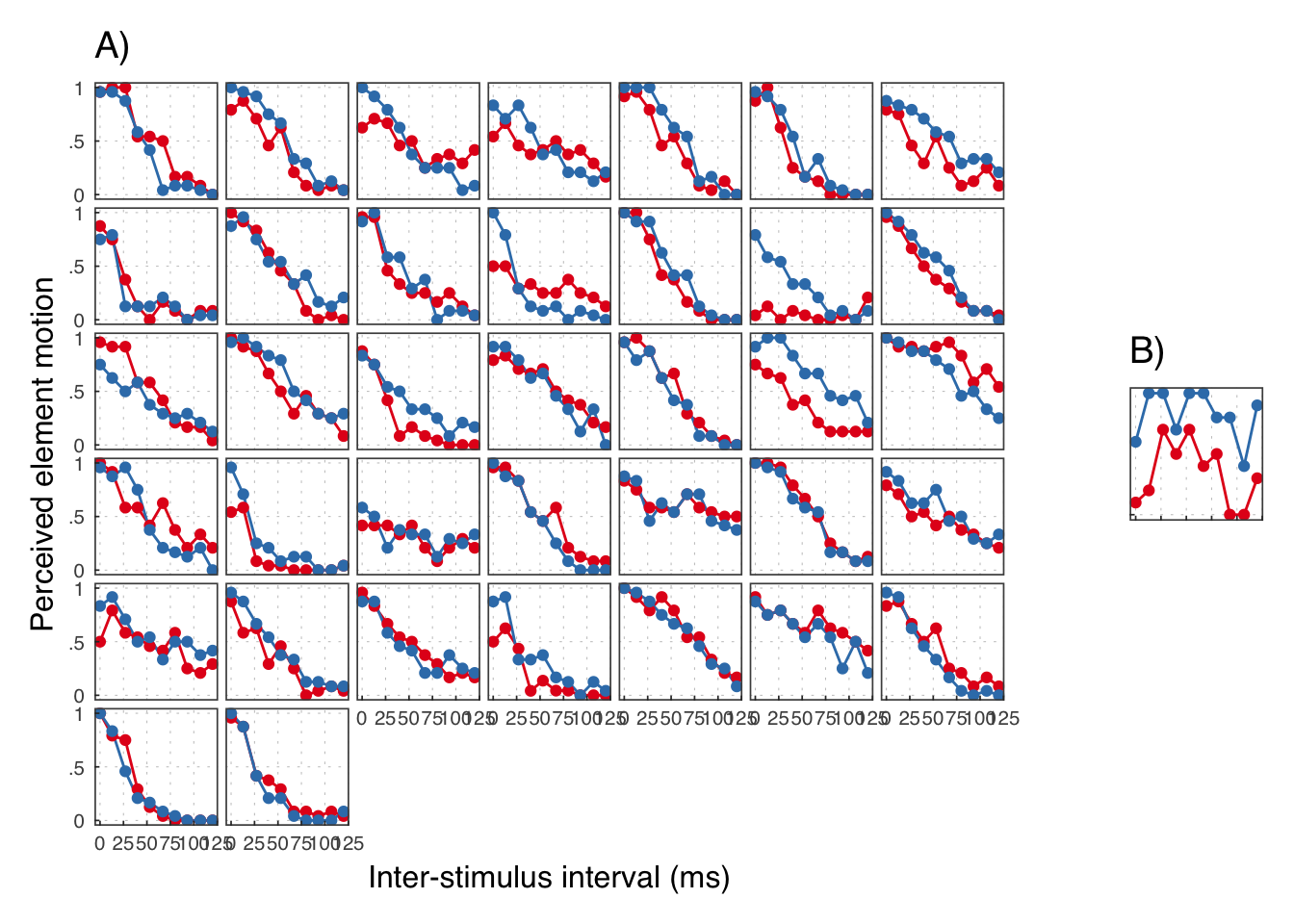
We reject the participants in B.
GLMM MLE estimation
exp3mle <- glmer(k/n ~ isic*condition + (isic*condition|id),
weights = n, family=binomial(link=logit),
data=exp3)summary(exp3mle)## Generalized linear mixed model fit by maximum likelihood (Laplace
## Approximation) [glmerMod]
## Family: binomial ( logit )
## Formula: k/n ~ isic * condition + (isic * condition | id)
## Data: exp3
## Weights: n
##
## AIC BIC logLik deviance df.resid
## 3375.7 3440.2 -1673.8 3347.7 726
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.9882 -0.6453 0.0293 0.7307 7.3409
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## id (Intercept) 0.97817 0.9890
## isic 0.04761 0.2182 0.19
## conditionvoluntary 0.44366 0.6661 -0.62 -0.01
## isic:conditionvoluntary 0.02776 0.1666 -0.08 -0.57 0.19
## Number of obs: 740, groups: id, 37
##
## Fixed effects:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -0.25511 0.16570 -1.540 0.1237
## isic -0.40656 0.03737 -10.878 <2e-16 ***
## conditionvoluntary 0.25607 0.11812 2.168 0.0302 *
## isic:conditionvoluntary -0.05164 0.03115 -1.658 0.0973 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) isic cndtnv
## isic 0.184
## cndtnvlntry -0.618 -0.023
## isc:cndtnvl -0.075 -0.576 0.164
## convergence code: 0
## Model failed to converge with max|grad| = 0.00147215 (tol = 0.001, component 1)GLMM Bayes estimation
exp3b <- brm(k | trials(n) ~ isic*condition + (isic*condition|id),
family=binomial(link=logit), prior = Priors,
data=exp3, cores = 4, iter = iters, warmup = warmups)## Inference for Stan model: binomial(logit) brms-model.
## 4 chains, each with iter=17500; warmup=5000; thin=1;
## post-warmup draws per chain=12500, total post-warmup draws=50000.
##
## mean se_mean sd 2.5% 97.5% n_eff Rhat
## b_Intercept -0.248 0.002 0.178 -0.598 0.101 6657 1
## b_isic -0.405 0.000 0.040 -0.486 -0.328 9487 1
## b_conditionvoluntary 0.253 0.001 0.127 0.002 0.503 10594 1
## b_isic:conditionvoluntary -0.055 0.000 0.033 -0.120 0.012 15592 1
##
## Samples were drawn using NUTS(diag_e) at Thu Feb 23 10:58:10 2017.
## For each parameter, n_eff is a crude measure of effective sample size,
## and Rhat is the potential scale reduction factor on split chains (at
## convergence, Rhat=1).# Posterior probability (one-sided p-value)
mean(posterior_samples(exp3b, pars = "b_cond") > 0) * 100## [1] 97.574## # A tibble: 3 × 4
## key m lwr upr
## <chr> <dbl> <dbl> <dbl>
## 1 Difference 6.281609 0.1284105 12.65075
## 2 Involuntary 43.905706 35.2922935 52.56462
## 3 Voluntary 50.187315 43.9534229 56.85072## # A tibble: 3 × 2
## key pp
## <chr> <dbl>
## 1 Difference 0.9772
## 2 Involuntary 1.0000
## 3 Voluntary 1.0000Experiment 4
( exp4 <- filter(illusion, exp == 4) )## # A tibble: 720 × 8
## exp id condition cond interval k n exclude
## <chr> <dbl> <chr> <dbl> <dbl> <dbl> <int> <lgl>
## 1 4 403 no_warning 0 0 21 24 FALSE
## 2 4 403 no_warning 0 13 21 24 FALSE
## 3 4 403 no_warning 0 27 16 24 FALSE
## 4 4 403 no_warning 0 40 13 24 FALSE
## 5 4 403 no_warning 0 53 17 24 FALSE
## 6 4 403 no_warning 0 67 8 24 FALSE
## 7 4 403 no_warning 0 80 12 24 FALSE
## 8 4 403 no_warning 0 93 7 24 FALSE
## 9 4 403 no_warning 0 107 5 24 FALSE
## 10 4 403 no_warning 0 120 1 24 FALSE
## # ... with 710 more rows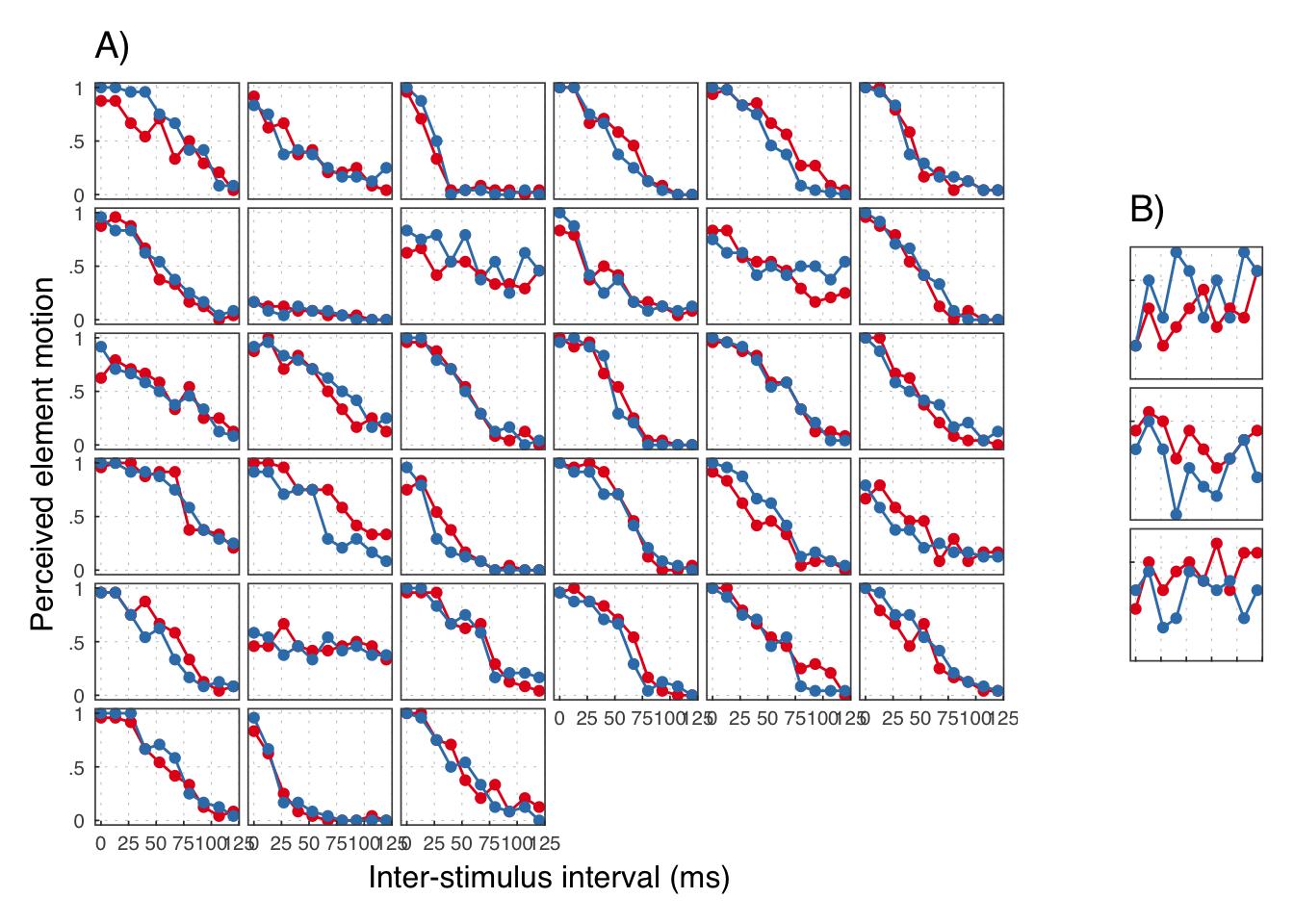
We reject the participants in B.
GLMM MLE estimation
exp4mle <- glmer(k/n ~ isic*condition + (isic*condition|id),
weights = n, family=binomial(link=logit),
data=exp4)summary(exp4mle)## Generalized linear mixed model fit by maximum likelihood (Laplace
## Approximation) [glmerMod]
## Family: binomial ( logit )
## Formula: k/n ~ isic * condition + (isic * condition | id)
## Data: exp4
## Weights: n
##
## AIC BIC logLik deviance df.resid
## 2815.3 2878.2 -1393.6 2787.3 646
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.6876 -0.6251 0.0345 0.7519 7.0223
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## id (Intercept) 0.959136 0.9794
## isic 0.038146 0.1953 0.02
## conditionwarning 0.137033 0.3702 -0.07 0.23
## isic:conditionwarning 0.008463 0.0920 0.19 0.15 0.06
## Number of obs: 660, groups: id, 33
##
## Fixed effects:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 0.03681 0.17399 0.212 0.832
## isic -0.49405 0.03601 -13.721 <2e-16 ***
## conditionwarning -0.02879 0.08109 -0.355 0.723
## isic:conditionwarning -0.02762 0.02347 -1.177 0.239
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) isic cndtnw
## isic 0.015
## condtnwrnng -0.138 0.178
## isc:cndtnwr 0.134 -0.065 0.037table(exp4$id)##
## 403 404 405 406 407 408 409 410 411 412 413 416 417 418 419 420 421 422
## 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20
## 423 424 425 426 428 429 430 431 432 433 434 435 436 437 438
## 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20GLMM Bayes estimation
exp4b <- brm(k | trials(n) ~ isic*condition + (isic*condition|id),
family=binomial(link=logit), prior = Priors,
data=exp4, cores = 4, iter = iters, warmup = warmups)## Inference for Stan model: binomial(logit) brms-model.
## 4 chains, each with iter=17500; warmup=5000; thin=1;
## post-warmup draws per chain=12500, total post-warmup draws=50000.
##
## mean se_mean sd 2.5% 97.5% n_eff Rhat
## b_Intercept 0.035 0.002 0.194 -0.346 0.414 6430 1
## b_isic -0.495 0.000 0.040 -0.575 -0.418 11964 1
## b_conditionwarning -0.026 0.001 0.089 -0.202 0.148 25845 1
## b_isic:conditionwarning -0.027 0.000 0.025 -0.077 0.021 33775 1
##
## Samples were drawn using NUTS(diag_e) at Thu Feb 23 11:01:35 2017.
## For each parameter, n_eff is a crude measure of effective sample size,
## and Rhat is the potential scale reduction factor on split chains (at
## convergence, Rhat=1).# Posterior probability (one-sided p-value)
mean(posterior_samples(exp4b, pars = "b_cond") > 0) * 100## [1] 38.34## # A tibble: 3 × 2
## key pp
## <chr> <dbl>
## 1 Difference 0.38266
## 2 No Warning 1.00000
## 3 Warning 1.00000## # A tibble: 3 × 4
## key m lwr upr
## <chr> <dbl> <dbl> <dbl>
## 1 Difference -0.4917079 -3.880532 2.976048
## 2 No Warning 50.7151923 42.981034 58.508879
## 3 Warning 50.2234844 42.574335 58.167847Combined analysis
We also compared the effect of condition across the Ternus experiments using an interaction term. That is, is the effect of condition greater in experiments 2 and 3, than in experiment 4?
( fulldata <- illusion %>% filter(exp > 1, !exclude) )## # A tibble: 2,060 × 8
## exp id condition cond interval k n exclude
## <chr> <dbl> <chr> <dbl> <dbl> <dbl> <int> <lgl>
## 1 2 202 involuntary 0 0 17 24 FALSE
## 2 2 202 involuntary 0 13 20 24 FALSE
## 3 2 202 involuntary 0 27 16 24 FALSE
## 4 2 202 involuntary 0 40 19 24 FALSE
## 5 2 202 involuntary 0 53 19 24 FALSE
## 6 2 202 involuntary 0 67 16 24 FALSE
## 7 2 202 involuntary 0 80 13 24 FALSE
## 8 2 202 involuntary 0 93 10 24 FALSE
## 9 2 202 involuntary 0 107 18 24 FALSE
## 10 2 202 involuntary 0 120 7 24 FALSE
## # ... with 2,050 more rowsfulldata$isic <- ( fulldata$interval - Cc ) / 10GLMM MLE Estimation
fullmle <- glmer(k/n ~ isic*cond + exp*isic + exp*cond +
(isic*cond|id),
weights=n, family=binomial(link=logit),
data=fulldata)## Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control
## $checkConv, : Model failed to converge with max|grad| = 0.0114011 (tol =
## 0.001, component 1)summary(fullmle)## Generalized linear mixed model fit by maximum likelihood (Laplace
## Approximation) [glmerMod]
## Family: binomial ( logit )
## Formula: k/n ~ isic * cond + exp * isic + exp * cond + (isic * cond |
## id)
## Data: fulldata
## Weights: n
##
## AIC BIC logLik deviance df.resid
## 9130.2 9242.8 -4545.1 9090.2 2040
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -3.6651 -0.6525 0.0077 0.7289 7.1248
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## id (Intercept) 0.93472 0.9668
## isic 0.04454 0.2110 0.27
## cond 0.27952 0.5287 -0.45 -0.07
## isic:cond 0.01699 0.1304 -0.05 -0.36 0.26
## Number of obs: 2060, groups: id, 103
##
## Fixed effects:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -0.41374 0.17236 -2.400 0.016377 *
## isic -0.48214 0.03644 -13.230 < 2e-16 ***
## cond 0.35137 0.10233 3.434 0.000595 ***
## exp3 0.15485 0.23977 0.646 0.518390
## exp4 0.44358 0.24301 1.825 0.067947 .
## isic:cond -0.02398 0.01595 -1.504 0.132697
## isic:exp3 0.06101 0.04922 1.240 0.215137
## isic:exp4 -0.01421 0.04996 -0.284 0.776073
## cond:exp3 -0.07537 0.14043 -0.537 0.591464
## cond:exp4 -0.37480 0.14341 -2.613 0.008962 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) isic cond exp3 exp4 isc:cn isc:x3 isc:x4 cnd:x3
## isic 0.267
## cond -0.450 -0.015
## exp3 -0.728 -0.188 0.317
## exp4 -0.708 -0.185 0.317 0.514
## isic:cond -0.022 -0.222 0.134 -0.018 0.004
## isic:exp3 -0.195 -0.705 -0.019 0.269 0.137 -0.038
## isic:exp4 -0.189 -0.687 -0.010 0.137 0.263 -0.006 0.514
## cond:exp3 0.332 -0.009 -0.724 -0.455 -0.234 -0.008 0.016 0.007
## cond:exp4 0.323 -0.005 -0.697 -0.235 -0.457 -0.020 0.009 0.017 0.511
## convergence code: 0
## Model failed to converge with max|grad| = 0.0114011 (tol = 0.001, component 1)GLMM Bayes estimation
fullb <- brm(k | trials(n) ~ isic*cond + exp*isic + exp*cond +
(isic*cond|id),
family=binomial(link=logit), prior = Priors,
data=fulldata, cores = 4, iter = iters, warmup = warmups,
save_ranef = FALSE) # Save space## Family: binomial(logit)
## Formula: k | trials(n) ~ isic * cond + exp * isic + exp * cond + (isic * cond | id)
## Data: fulldata (Number of observations: 2060)
## Samples: 4 chains, each with iter = 17500; warmup = 5000; thin = 1;
## total post-warmup samples = 50000
## WAIC: Not computed
##
## Group-Level Effects:
## ~id (Number of levels: 103)
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample
## sd(Intercept) 1.00 0.08 0.86 1.17 11235
## sd(isic) 0.22 0.02 0.19 0.26 12578
## sd(cond) 0.55 0.05 0.46 0.66 17779
## sd(isic:cond) 0.13 0.01 0.11 0.16 20396
## cor(Intercept,isic) 0.26 0.10 0.05 0.45 9725
## cor(Intercept,cond) -0.43 0.10 -0.60 -0.22 21907
## cor(isic,cond) -0.05 0.12 -0.28 0.18 14458
## cor(Intercept,isic:cond) -0.03 0.13 -0.28 0.22 19748
## cor(isic,isic:cond) -0.33 0.11 -0.54 -0.09 20461
## cor(cond,isic:cond) 0.24 0.13 -0.02 0.48 15893
## Rhat
## sd(Intercept) 1
## sd(isic) 1
## sd(cond) 1
## sd(isic:cond) 1
## cor(Intercept,isic) 1
## cor(Intercept,cond) 1
## cor(isic,cond) 1
## cor(Intercept,isic:cond) 1
## cor(isic,isic:cond) 1
## cor(cond,isic:cond) 1
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
## Intercept -0.41 0.18 -0.77 -0.06 4998 1
## isic -0.48 0.04 -0.55 -0.41 8096 1
## cond 0.35 0.11 0.15 0.56 10047 1
## exp3 0.16 0.25 -0.33 0.64 4592 1
## exp4 0.45 0.25 -0.05 0.94 4750 1
## isic:cond -0.03 0.02 -0.06 0.01 23692 1
## isic:exp3 0.06 0.05 -0.04 0.16 7519 1
## isic:exp4 -0.02 0.05 -0.12 0.09 8059 1
## cond:exp3 -0.08 0.14 -0.36 0.21 9778 1
## cond:exp4 -0.38 0.15 -0.67 -0.08 10662 1
##
## Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
## is a crude measure of effective sample size, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Posterior probability that effect of condition (i.e. warning condition effect is smaller than voluntary action condition effect).
# Posterior probability (one-sided p-value)
mean(posterior_samples(fullb, pars = "b_cond:exp4") < 0) * 100## [1] 99.4Compare effect of condition across the three Ternus experiments:
x <- as.data.frame(fullb, pars = "b_cond") %>%
as_tibble() %>%
transmute(cond_exp2 = b_cond,
cond_exp3 = b_cond + b_cond.exp3,
cond_exp4 = b_cond + b_cond.exp4,
cond_2v3 = b_cond.exp3,
cond_2v4 = b_cond.exp4,
cond_3v4 = cond_exp4 - cond_exp3)
x %>%
gather() %>%
group_by(key) %>%
summarize(bhat = mean(value),
`2.5%ile` = quantile(value, probs = 0.025),
`97.5%ile` = quantile(value, probs = 0.975))## # A tibble: 6 × 4
## key bhat `2.5%ile` `97.5%ile`
## <chr> <dbl> <dbl> <dbl>
## 1 cond_2v3 -0.07702028 -0.36268402 0.20589888
## 2 cond_2v4 -0.37716975 -0.67038813 -0.08425850
## 3 cond_3v4 -0.30014947 -0.58698900 -0.01374827
## 4 cond_exp2 0.35233824 0.14620466 0.56059514
## 5 cond_exp3 0.27531796 0.07914614 0.47232450
## 6 cond_exp4 -0.02483151 -0.23728027 0.18591864